| Instructions | Output format | Data | Software |
EffectorP: predicting fungal and oomycete effector proteins from secretomes using machine learning
Introduction
What is EffectorP?
Many fungi and oomycete species are devastating plant pathogens.
These eukaryotic filamentous pathogens secrete effector proteins to facilitate plant infection.
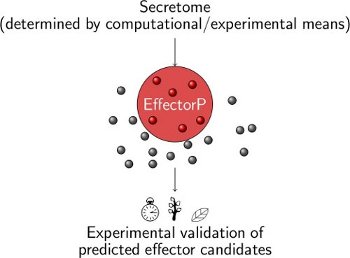
EffectorP is a machine learning method for fungal and oomycete effector prediction in secretomes.
Fungi and oomycete pathogens have diverse infection strategies and their effectors generally do not share sequence homology.
However, they occupy similar host environments, either the plant apoplast or plant cytoplasm,
and may therefore share some unifying properties based on the requirements of these host compartments.
EffectorP 3.0 exploits this biological signal and uses two machine learning models:
one trained on apoplastic effectors and one trained on cytoplasmic effectors.
What is EffectorP not?
EffectorP has been trained to distinguish secreted proteins from secreted effectors in plant-pathogenic fungi and oomycetes.
It has not been trained on data from bacteria, nematodes or insects and will not accurately predict effectors in these species.
EffectorP is also not a tool for secretome prediction. EffectorP 3.0 recognizes a cytoplasmic signal also in non-secreted, intracellular proteins.
It is essential to first use tools such as SignalP, Phobius and TMHMM to predict if a protein is likely to be secreted.
Which version of EffectorP should I run?
EffectorP 3.0: for both fungal and oomycete effector prediction. Distinguishes between apoplastic and cytoplasmic effectors.
EffectorP-fungi 3.0: for fungal effector prediction. Distinguishes between apoplastic and cytoplasmic effectors. Might be slightly less accurate than EffectorP 3.0.
EffectorP 2.0: for fungal effector prediction. Less accurate than EffectorP 3.0.
EffectorP 1.0: for fungal effector prediction. Less accurate than EffectorP 3.0.
Submission
To run EffectorP, please submit your secreted fungal or oomycete proteins of interest below. For online submission, the maximum number of protein sequences that can be submitted is 4000. If you want to run EffectorP on your local machine, you can download the current versions here.
i
Citation for EffectorP 3.0
Citation for EffectorP 2.0
Citation for EffectorP 1.0
Contact
For comments, suggestions and reporting bugs please contact jana.sperschneider@csiro.au
